A series of discoveries and technical advancements over the past decade has spawned a new field called RNA epigenetics or epitranscriptomics, all the biochemical modifications of the RNA (the transcriptome) within a cell, and the proteins that write, erase, and read these modifications. In recent years, studies implicating epitranscriptomic proteins in cancer have led to the launch of several biotech companies, e.g., Storm Therapeutics, Accent Therapeutics, Gotham Therapeutics, Twentyeight-Seven Therapeutics, etc. dedicated to drugging these proteins. There was a similar reaction to epigenetics more than a decade ago, when it became clear that chemical modifications regulating genes are frequently out of whack in cancer. Companies rushed to develop drugs against proteins responsible for making, removing, and recognizing chemical modifications on genes – often referred to as the writer, eraser, and reader proteins. With the discovery of parallel writer, eraser, and reader proteins working on RNA, epitranscriptomics is looking like a promising, untapped area for drug discovery.
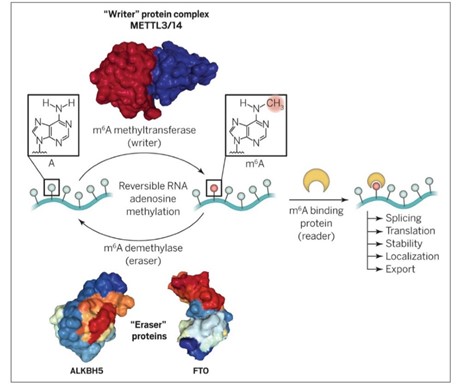
Innovations in epitranscriptomics have resulted in the identification of more than 160 RNA modifications to date. These modifications happen to many types of cellular RNA, including but not limited to, ribosomal RNA (rRNA), transfer RNA (tRNA), messenger RNA (mRNA), and small nuclear RNA (snRNA). The most common and well-understood mRNA modification at present is N6-Methyladenosine (m6A), which has been observed to occur an average of three times in every mRNA molecule. Currently, work is focused on determining the types of and location of RNA modifications, determining if these modifications have function, and if so, what is their mechanism of action. One function that has been elucidated involves the enzyme adenosine deaminase (ADAR), which acts on RNA. ADAR affects a series of cellular processes, including alternative splicing, microRNAs, the innate immune system, and leads to protein recoding especially for important receptors in the central nervous system. Proteins that recognize and bind to m6A are known as "readers". However, some of the mRNA modifications prevent the binding of some RNA binding proteins; these are called "anti-readers". The "writers" and "erasers" of the m6A mark are mostly located in nuclear speckles (subnuclear structures enriched in pre-mRNA splicing factors), where mRNA is processed and stored. The m6A mark is added by an m6A methyltransferase complex post-transcriptionally. This "writer" complex is composed of METTL3, METTL14, Wilms tumor 1-associated protein (WTAP), KIAA1429 and RBM15.
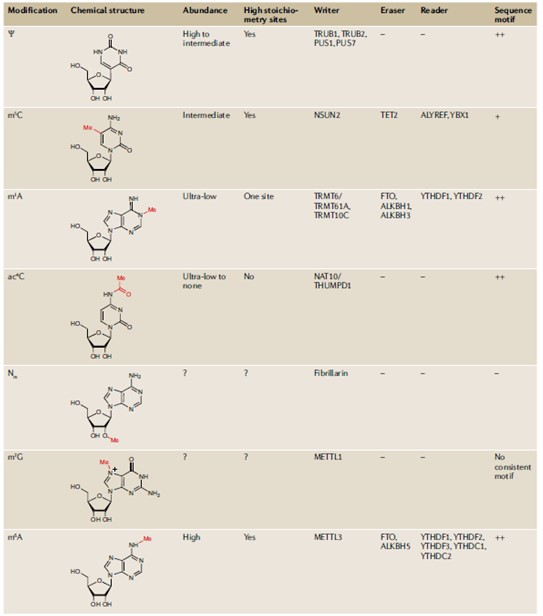
Some other RNA modifications include pseudouridine (Ψ), 5- methylcytidine (m5C), N1-methyladenosine (m1A), N4- acetylcytidine (ac4C), ribose methylations (Nm), and N7- methylguanosine (m7G). The table below shows the abundance, stoichiometry, machinery, and consensus motifs of these RNA modifications.
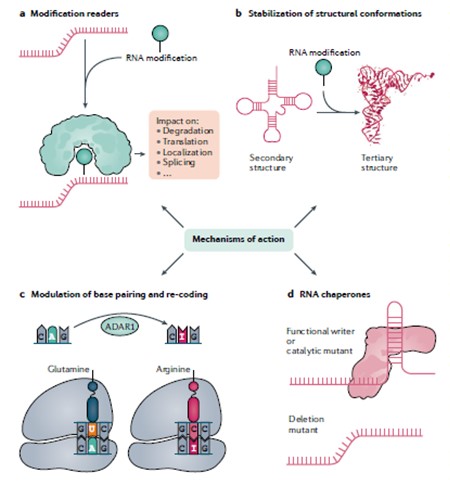
RNA modifications can act through various mechanisms (figure below): a) A specialized protein (‘reader’) specifically binds the modification and triggers different molecular phenotypes downstream. b) The chemical properties of the modification help to stabilize a specific RNA structure. c) The modification leads to altered Watson-Crick interactions, which can lead, for example, to a difference in the decoding of the modified base by the translational machinery. This is illustrated for an adenosine base that can be edited into inosine, which will be decoded as a guanosine. d) The modification ‘writer’ serves as a chaperone for proper folding of the RNA. The modification itself is not strictly required for such chaperoning activity and, therefore, catalytic mutants lacking the modification will allow similar folding activity as the wild type but different than in a full deletion of the modification writer.
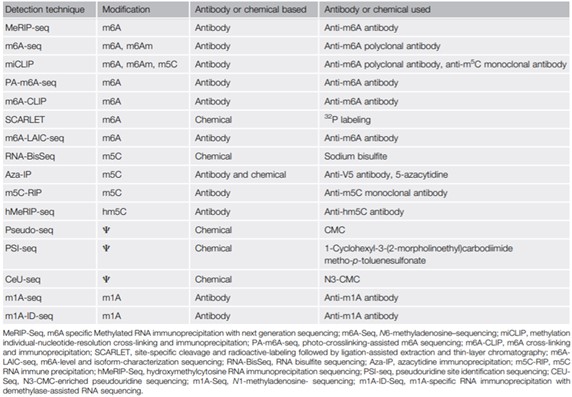
Currently, the detection techniques used for transcriptome-wide identification of chemical modifications mainly depend on chemical and antibody-based detection methods followed by sequencing analysis (table below). However, fourth-generation sequencing technologies such as Oxford Nanopore Technologies (ONT), which promise to provide long read sequencing of the native full-length RNA transcripts, open new frontiers for RNA modification detection, like the developments made on DNA modification detection on native DNA fragments in the recent past.
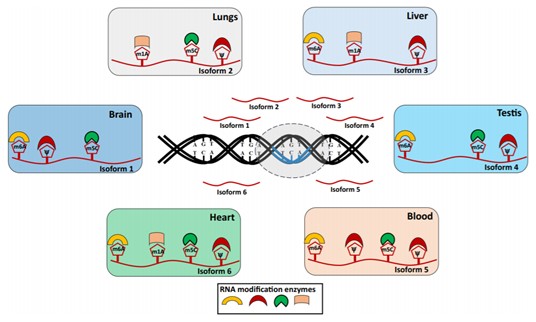
Although recent developments in epitranscriptomics approaches have enabled the transcriptome-wide mapping of several RNA modifications, several limitations still exist. Highly sensitive and accurate approaches are needed to identify, quantify, and monitor RNA modifications that occur at low abundance across classes of RNAs (rRNAs, tRNAs, snoRNAs, microRNAs, mRNAs, and long ncRNAs) on individual transcript isoforms to uncover the role of epitranscriptome in modulating the spatiotemporal cellular regulatory networks (figure below). Although a combination of transcript isoforms of a gene is expressed in a tissue, their dynamic tissue/cell-type-specific regulation by different modification enzymes (writers, readers, and erasers) via the epitranscriptome code determines the differential post-transcriptional regulatory fate of an RNA molecule resulting from a locus. Such combinatorial epitranscriptome marks specific to an RNA transcript originating from a genic locus can dictate its splicing, stability, and localization as well as its translation status, thereby providing a precise cell-type-specific spatiotemporal context for regulation.
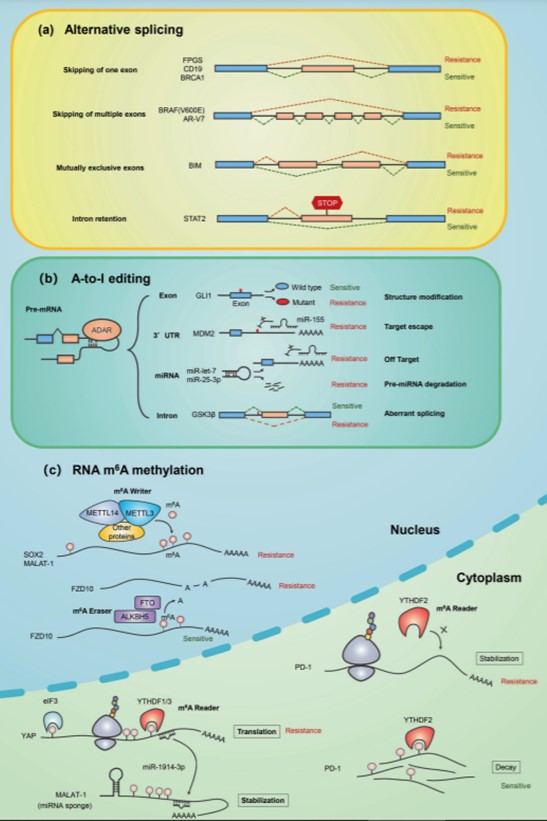
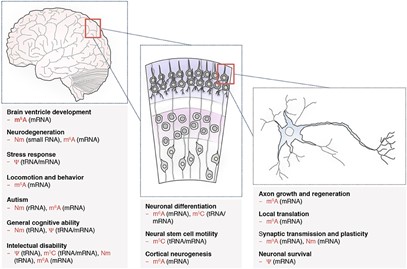
Alterations in the pathways leading to the addition of the removal of the m6A mark result in impaired gene expression and cellular function, which can lead to disease. Normal m6A levels are altered in several cancers. The figures below show RNA modifications in cancer drug resistance and those implicated in various neuronal processes. Reduced m6A levels due to down regulation of METTL3 and/or METTL14 lead to the activation of several oncogenes. All things considered, "writers" and "erasers" of the m6A mark may be good potential drug targets in cancer therapy. Metabolic disorders are also affected by the m6A mark due to the role of FTO (fat mass and obesity-associated gene).
Many see RNA editing as an important alternative to DNA editing using techniques such as CRISPR. CRISPR technology is improving, but DNA editing can cause unwanted mutations in other parts of the genome — ‘off-target effects’ — which might create new problems. But since ADARs are much less efficient than CRISPR, they could be less useful for making genetically modified plants and animals. As a research tool, it is very limiting. Another major disadvantage is that ADARs can make only a few kinds of change to RNA; ADARs are more like an overwrite function that changes letters chemically, without breaking the RNA molecule’s ‘backbone’. Although this process is less likely to cause unintended mutations, it limits the enzymes to making specific changes — adenosine to inosine in the case of ADARs, and cytosine to uridine by a set of enzymes called APOBECs.
Several other hurdles remain. Current technologies can alter RNA sequences in only a few limited ways and getting the system to work as intended in the human body will prove challenging. Still, researchers hope that new technologies, such as protein engineering, and improved methods for delivering RNA to cells can help to overcome these limitations. Despite its early stage, researchers and biotech investors are excited about the wide potential of RNA editing. For many years, this was a backwater, and suddenly there is a company popping up every two weeks. Numerous start-ups and established DNA-editing firms have announced their intention to move into RNA. The challenge for industry is to work out the best way to get the guide RNAs into the cell without triggering an immune reaction or causing the cell to degrade them. This could include making strategic chemical modifications to the engineered RNAs that stabilize them or embedding them in a nanoparticle or virus that can sneak into cells.